Introduction
Diagnostic tools which screen the binding interactions of a protein target against a library display of biomolecular probes to identify which probe molecules which bind the target are central to cell proteomic studies, to efforts at enzyme or antibody discovery, and are useful as biosensors for environmental surveillance and food monitoring, and biodiagnostic assays for detecting disease markers. For screening a target against a large number of probes traditional methods include using well plates, dispensing a different probe (shown below as antibodies) in each well and spotting probes on a surface. The binding of the target to the surface probes is detected by fluorescence, either by directly labeling the target or undertaking an enzyme linked sandwich assay. Alternatively, probes are conjugated to the surface of microbeads, and and flow cytometry is used to detect fluorescently labelled targets.
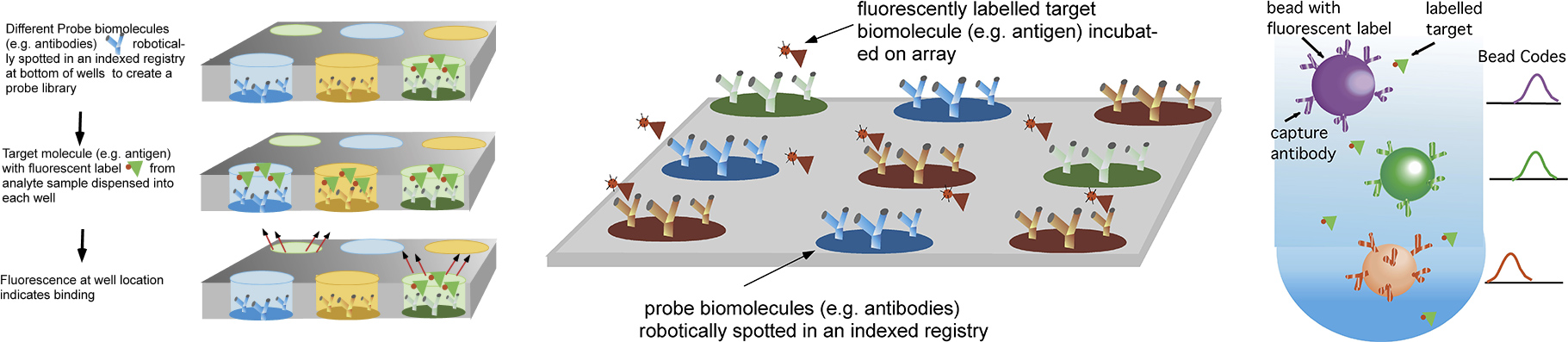
Methods for screening the binding interactions of a target with a probe library sequestered in wells, spotted on a surface or conjugated to microbeads for flow cytometry analysis
Our Research
Our research focuses on microfluidic “lab on a chip’’ designs for screening binding interactions in which the probe molecules are hosted on microbeads (as in flow cytometry) but are arrayed in a microfluidic channel. The ultraminiaturized microfluidic design allows for portability (point of care testing), high sensitivity and reduced consumption of reagents and target. To screen a target against a library of potential binding partners, analyte containing the target is flowed over the microbead array assembly, and the binding of the target to the surface probes is detected by fluorescence, either by directly labeling the target or undertaking an enzyme linked sandwich assay
Our researchstudies two designs for forming microbead arrays: In one, the beads are captured in an array of wells formed at the bottom of the microfluidic cell, and in the second the microbeads are captured in “V” shaped traps. In both designs, the microbeads are streamed through the trapping course and captured. Only one microbead is retained per trap or well. Sequential capture of microbeads, each with different probes on their surfaces, allows for multiplexed arrays displaying microbeads.
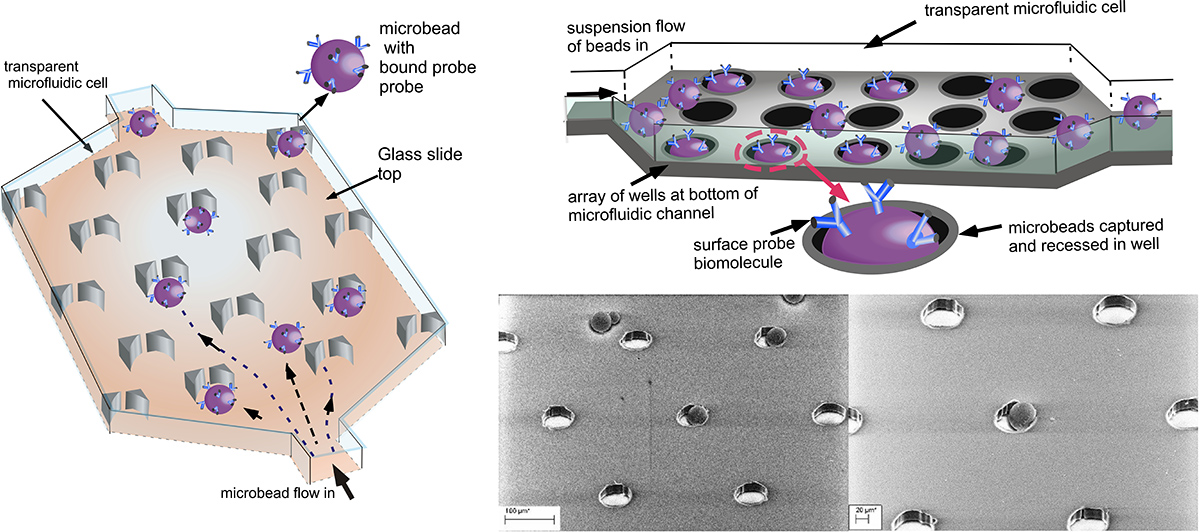
Using these designs, we have studied the mass transfer of the binding of the target to the surface probes, obtaining numerical solutions for the transport and comparing to experiments with fluorescently labelled Neutravidin as the protein target, and biot displayed on the microbead surface.


First two figures: Binding of fluorescently labelled Neutravidin to microbeads with surface avidin as a function of time. Movie: time lapse binding of labelled Neutravidin to biotinylated beads trapped in wells.
We have also studied the sequestering of cell membrane receptors as probes on the surface of the microbeads. Screening the interactions of a target with an array of these receptors is important in drug discovery – where most drugs are developed to target surface membrane receptors to exert a therapeutic effect. Libraries of surface receptors are also important in the detection of bacteria; pathogens, as many pathogens bind to receptors on the surface of biological cells before inserting themselves into the cell. Pathogens bind with high selectivity and affinity, and therefore recepetor arrays can be used to detect the pathogen in an analyte solution.
Membrane receptors require their native bilayer environment to retain their selectivity and binding affinity, and this complicates displaying them in a microarray platform. Here the membrane probes/receptors are first incorporated in supported lipid bilayers formed around glass microbeads (lipobeads), and the microbeads themselves are then arrayed on a surface by hydrodynamic capture in the microfluidic obstacle course of traps. We have illustrated how this approach can be used to assay for cholera toxin which binds to the surface of cells by complexing to a ganglioside, its binding partner, on the cell surface.
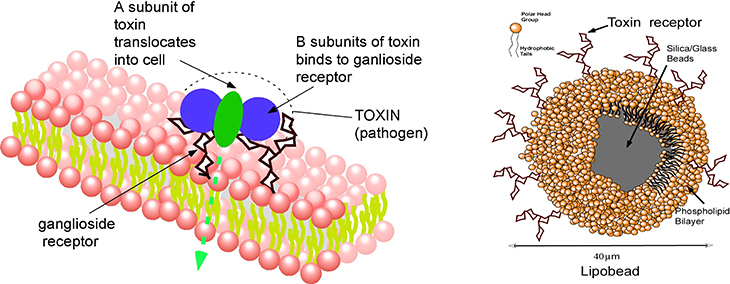
Detection of cholera toxin by using lipobeads displaying the cell membrane ganglioside that binds to unit of the toxin. The movie (cholera2.mov) shows the fluorescence of the labelled toxin as a function of time as it binds from solution to the lipobeads arrayed in traps.
